|
Platinum
Tutorial 6: Step 4 Run SLAM
Associations in Data
Sub-Linear Association Mining (SLAMô) is a method for finding associations in discrete data. An association is a set of variables (genes) and values which occur together in a dataset at a rate higher than that expected by chance. For instance, it might happen that in kidney tissue repression of gene A results in the up-regulation of genes B and C, and down-regulation of gene Q. In this case, we would expect to find an association in the dataset like this:
Kidney Tissue: Gene A: low, gene B: high, gene C: high, gene Q: low.
Note: this says nothing about how B, C, and Q are regulated when A is not repressed, or when a different tissue is being considered.
Such sets of variables have several potential uses. In GeneLinkerô, they are used to identify key sets of genes which might be predictive of a given sample classification. This use, called feature selection, is vital to making predictions because of the enormous number of genes in a microarray experiment which are typically not connected to the class of interest.
The SLAMô Parameters
Imagine you are searching for a book in a library, and you know it's Dewey Decimal number. One way you could find it would be to start at 100.00 and walk along the shelves until you get to the number of your book. This is not very efficient. Instead, you might walk around at random and glance at numbers now and then, making a random sampling of what books are near you at any given time. This is a surprisingly efficient strategy, and SLAMô uses something like it to find associations in gene expression data.
Two of the parameters in the dialog above relate to SLAMô's random sampling behavior. One is the Number of Iterations. This is the number of random subsets of your data SLAMô uses to find associations. The higher the number of iterations, the more and better associations will be found, but the longer the algorithm will take to run.
The second parameter is the Random Seed. This controls the sequence of random numbers that are used by the algorithm to select subsets. If the seed is set to the same value, and SLAMô is run again, it will produce identical results. Running SLAMô on the same data with different random seeds will produce similar, but not identical, results, because slightly different subsets will have been selected from the data.
The Representative Variable is the variable you want to classify on. Datasets may have several variables associated with them (cancer type, tissue type, gender, etc.) and you can use SLAMô to search for features that discriminate between values of any variable.
Support is the number of subsets an association must appear in before it is considered significant. Associations with less than the minimum support will not be reported.
Matthews Number is a measure of how good an association is at discriminating between classes. Perfect discrimination is represented by a Matthews number of 1. Useful values are typically between 0.5 and 0.7.
Run SLAMô on the Discretized Data
1. If the newly created Discretized: 3 bins/gene | quantile dataset in the Experiments navigator is not highlighted, click it.
2. Click the SLAMô
toolbar icon ![]() , or select SLAM
from the Predict menu, or right-click
the item and select SLAM from
the shortcut menu. The SLAMô parameters
dialog is displayed.
, or select SLAM
from the Predict menu, or right-click
the item and select SLAM from
the shortcut menu. The SLAMô parameters
dialog is displayed.
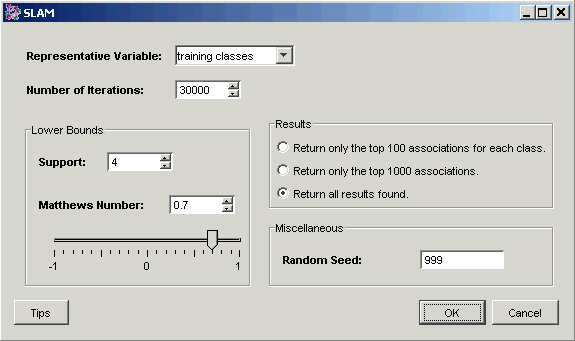
3. Set the dialog parameters.
|
Parameter |
Setting |
|
Representative Variable |
training classes |
|
Number of Iterations |
30000 |
|
Support |
4 |
|
Matthews Number |
0.7 |
|
Results |
Return all results found. |
|
Random Seed |
999 (see Note below) |
4. Click OK. The SLAMô operation is performed. This may take fifteen minutes or so, on an IBM box as described in the System Specification. Upon successful completion, a new item (SLAM) is added under the Discretization item in the Experiments navigator.
If you have automatic visualizations enabled in your user preferences, the SLAM Association Viewer is displayed.
Note on Use of the Random Seed Parameter
In normal use, setting the random seed is neither necessary nor recommended. In a tutorial you set the random seed to a consistent value so that you will obtain precisely the same results as we depict and discuss, which makes the tutorial easier to understand. When you are not following a tutorial, you should generally not adjust the random seed at all.
In SLAMô, the random seed can be thought of as prescribing the starting point for the search for associations. If SLAMô is allowed to run long enough, it will find all of an enormous set of associations which inhabit any given dataset, but the smaller you set the number of iterations, the greater will be the effect of the random seed. Conversely, the random seed matters less and less as the number of iterations grows greater. It is usually better to set the iteration number high and let SLAMô run overnight than to do repeated runs with different random seeds.

