|
Changing Your User Preferences
Overview
This facility allows GeneLinkerô to remember your preferences from one session to the next.
Actions
1. Select Preferences from the Tools menu. The User Preferences dialog is displayed.
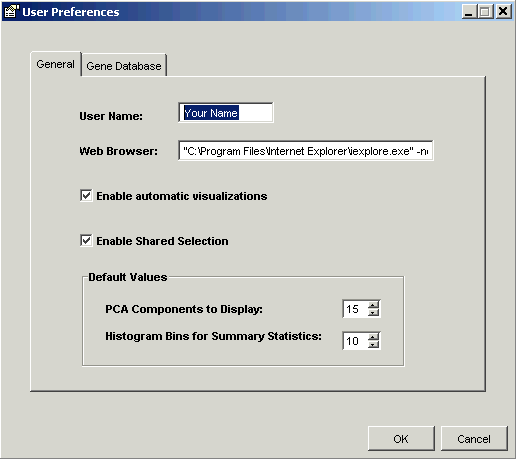
2. Click the General tab to display the general preferences pane.
3. Set the parameters.
|
Element |
Description |
|
User Name |
Your user identifier that appears in annotations and reports containing annotations. |
|
Web Browser |
The path to your preferred HTML browser. |
|
Enable automatic visualizations |
If this checkbox is checked then whenever any analysis experiment (any experiment other than data import, normalization, filtering, or missing value estimation) completes, the default visualization that is associated with the experiment will be opened automatically as soon as the experiment completes. By default this preference is checked. |
|
Enable Shared Selection |
If this checkbox is checked, items such as genes and samples that are selected in one visualization will also be highlighted in other visualizations. By default this preference is checked. |
|
PCA Components to Display |
The default number of principal components to display in a loadings line plot or loadings color matrix plot (display only - does not affect the calculation). |
|
Histogram Bins for Summary Statistics |
The default number of bins for the Summary Statistics chart. |
4. Click the Gene Database tab to display the gene database pane.
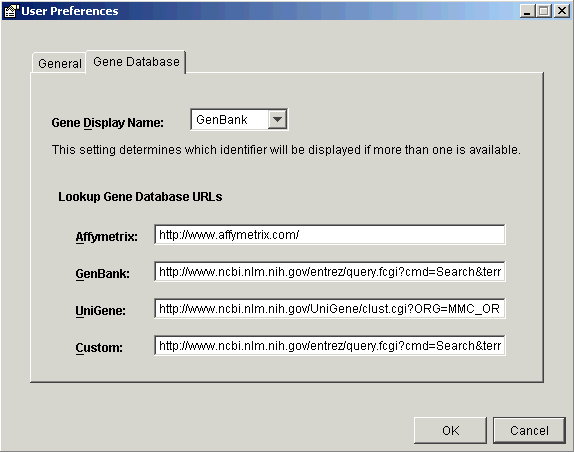
5. Set the parameters.
|
Element |
Description |
|
Gene Display Name |
The default type of gene identifier used for display. |
|
Lookup Gene Database URL: Affymetrix |
Database URL for looking up a gene with an Affymetrix gene identifier. See Affymetrix URL Format below. |
|
Lookup Gene Database URL: GenBank |
Database URL for looking up a gene with a GenBank gene identifier. See GenBank URL Format below. |
|
Lookup Gene Database URL: UniGene |
Database URL for looking up a gene with a UniGene gene identifier. See UniGene URL Format below. |
|
Lookup Gene Database URL: Custom |
The URL used to access another gene database. Use the correct URL format for the database you are accessing. |
6. Click OK to save changes to the settings or click Cancel to keep the previous values.
For more information about forming query strings, refer to Linking to PubMed and other Entrez databases: http://www.ncbi.nlm.nih.gov/entrez/query/static/linking.html
|
Affymetrix URL Format: |
https://www.netaffx.com/index2.jsp
|
GenBank URL Format: |
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Search&term=PPSI_ID&db=Nucleotide&doptcmdl=GenBank
Note the use of the term PPSI_ID. This term must appear in the URL. The application will replace this term with the identifier of a gene.
For example, if the gene being queried has the identifier AF098020, then the application will use the following URL to obtain information about that gene:
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Search&term=AF098020&db=Nucleotide&doptcmdl=GenBank
|
UniGene URL Format: |
http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=PPSI_ORGANISM&CID=PPSI_ID
Note: the use of the terms PPSI_ORGANISM and PPSI_ID. These terms must appear in the URL. The application will replace these terms with the appropriate components of a UniGene gene identifier.
For example, if the gene being queried has the UniGene identifier Ht.9573, then the application will use the following URL to obtain information about that gene:
http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Ht&CID=9573
Related Topics:

