|
Performing K-Means Clustering
Overview
K-Means clustering generates a specific number of disjoint, flat (non-hierarchical) clusters. It is well suited to generating globular clusters.
For further details, see Overview of K-Means Clustering.
Actions
1. Click a complete dataset in the Experiments navigator. The item is highlighted.
2. Click the Partitional
Clustering toolbar icon ![]() , or select Partitional
Clustering from the Clustering
menu, or right-click the item and select Partitional
Clustering from the shortcut menu. The Partitional
Clustering parameters dialog is displayed.
, or select Partitional
Clustering from the Clustering
menu, or right-click the item and select Partitional
Clustering from the shortcut menu. The Partitional
Clustering parameters dialog is displayed.
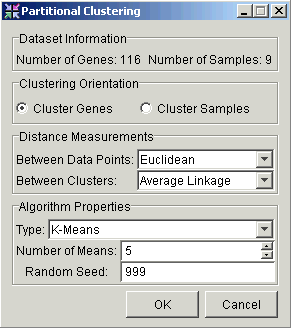
3. Set the parameters.
|
Parameter |
Description |
|
Clustering Orientation |
Cluster by Genes or by Samples. |
|
Distance Measurement Between Data Points |
Type of distance measurement to use to determine how close two data points are to each other. |
|
Distance Measurement Between Clusters |
Type of distance measurement to use to determine how close two clusters are to each other. |
|
Type |
Set this parameter to K-Means. |
|
Number of Means |
This value specifies the number of clusters the algorithm forms. The value must be greater than or equal to 2, and less than or equal to the number of clusterable items (genes or samples) in the selected dataset. |
|
Random Seed |
The seed value for the random number generator. In normal use, setting the random seed is neither necessary nor recommended. On occasion, you may need to determine whether a certain variation in results is due to the random element, or some other cause. For this reason, you are able to set the random seed to a fixed value, thus controlling that source of variation. |
4. Click OK. The Experiment Progress dialog is displayed. It is dynamically updated as the K-Means Clustering operation is performed. To cancel the K-Means Clustering operation, click the Cancel button.
Upon successful completion, a new item is added under the original item in the Experiments navigator.
Related Topics:

