|
Platinum
IBIS Search
Overview
The IBIS search examines all of the genes (or gene pairs) in a dataset as predictors for a target variable. If you already know which gene or gene pair you would like to use to create an IBIS classifier, you do not need to perform an IBIS search. Please see Create IBIS Classifier Using a Gene or Gene Pair.
The IBIS search process creates proto-classifiers using the specified parameters and generates accuracy and MSE statistics for each. An item is added to the Experiments navigator which contains a list of the proto-classifiers and their associated statistics. At the end of the search process, no true classifiers exist, only the information about them and how to produce them, hence the term proto-classifier.
There are three models available for creating classifiers: Linear Discriminant Analysis (LDA), Quadratic Discriminant Analysis (QDA), and Uniform/Gaussian Discriminant Analysis (UGDA). In general, it is best to start by creating classifiers using LDA and single genes. Only if the accuracy and MSE values are unsatisfactory should you try QDA/UGDA as well as gene pairs.
For a single gene search, one proto-classifier is created for each gene in the dataset (to a maximum of 10000). For a gene pair search, one proto-classifier is created for each pair of genes in the dataset (to a maximum of 1000). Generating a list of IBIS proto-classifiers for gene pairs takes much longer than for single genes. It is recommended that you filter your dataset before performing the search to remove any genes that are not relevant to the search.
Actions
1. Click a complete dataset with variable information
item (![]() dataset name) in the Experiments
navigator. The item is highlighted.
dataset name) in the Experiments
navigator. The item is highlighted.
2. Select IBIS Classifier Search from the Predict menu, or right-click the item and select IBIS Classifier Search from the shortcut menu. The IBIS Classifier Search dialog is displayed.
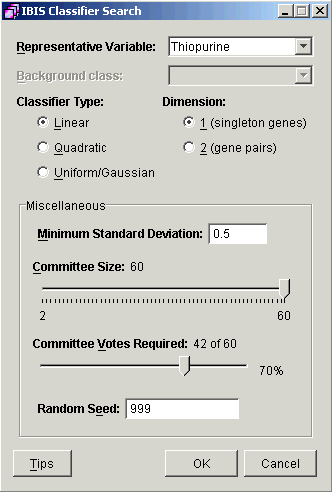
3. Set parameters.
|
Parameter |
Description |
|
Representative Variable |
It cannot contain the class 'unknown' and it must have at least two classes with a minimum of three observations (samples) for each class. |
|
Background Class (UGDA only) |
Representative variable class to be used as the background reference. Suggestion: select the variable value with the highest frequency in the training data. |
|
Classifier Type |
Select Linear, Quadratic, or Uniform/Gaussian. |
|
Classifier Dimension |
Dimension of the resultant classifier (1 or 2 genes). |
|
Minimum Standard Deviation |
Use this value to capture your estimate of the error in the data measurements. If the value is too small, degenerate non-useful patterns may be created. If the value is too large, you may miss important patterns due to over-smoothing the classifier. As the name suggests, an appropriate value would be the smallest standard deviation of the expression of any gene/sample pair over a number of replicate measurements. For full details on this parameter, see Tutorial 7: Appendix. |
|
Committee Size |
Number of individual classifiers in the classifier. |
|
Committee Votes Required |
Threshold for classifier to make a prediction. |
|
Random Seed |
An initial value for the random number generator. In IBIS, randomization is only used in cross-validation and the committee structure, that is, in designating training and internal validation samples. |
4. Click OK. The IBIS search is performed and upon successful completion, a new IBIS Search item is added under the original dataset in the Experiments navigator.
Visualization
The IBIS Search Results Viewer can be used to examine the results of the IBIS search operation.
Related Topics:
Create IBIS Classifier From IBIS Search Results

