Overview
GeneLinker™ scripts are generated by selecting an experiment and choosing one of the Generate Script options from the Tools menu. Please see Generating Scripts for more detail on this process.
To run a script, an experiment must be selected in the Experiments navigator, and then a script file selected using the Run Script item in the Tools menu. The script file may be a workflow or single experiment script.
The experiments described in the script must be appropriate to the selected data. For example, if a script that includes Filtering by Reliability Measures is applied to a dataset that does not include reliability measures, the script will fail, presenting an error dialog.
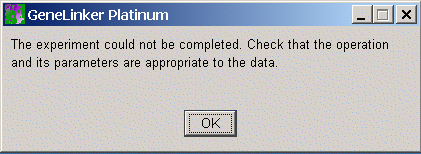
Although many problems with script parameters will result in informative error messages, in some cases the only result will be a generic error dialog:

In this case, it should be possible to determine the cause of the problem by checking the parameters of the experiments used to generate the script.
For workflow scripts, each step in the script will be run until the script reaches completion, or until a step fails, at which point the script will terminate. The only exception to this rule is Missing Value Estimation experiments: if a workflow script contains a Missing Value Estimation step but there are no missing values in the table it is applied to, the step will silently be skipped. Note that for Missing Value Estimation scripts the fraction of missing values that will trigger gene removal is saved in the script, not the absolute number. So if a script was generated from a dataset with 36 samples with 18 missing values as the threshold for gene removal, if it is run on a dataset with 100 samples the threshold for gene removal will be 50 missing values.
The most frequent problems with scripts come from the following:
Careful attention to the messages in the error dialogs will aid in understanding the cause of the problem.
Currently there is no way to examine or edit the parameters in a script once it has been generated. This capability will be added in a future version of GeneLinker™.
While a script is running, the application will be very busy updating the Experiments navigator and running default visualizations. On rare occasions this will result in slight defects in the appearance of the application which may safely be ignored.
Actions
1. Click an item in the Experiments navigator. The item is highlighted.
2. Select Run Script from the Tools menu. The Open dialog is displayed.
3. Navigate to the folder where the file is to be opened.
4. Select the script file to run.
5. Click Open. The script will load and run.
Related Topics: