|
Platinum
Tutorial 9: Introduction
This tutorial continues to explore data mining and prediction. You will use the committee of Support Vector Machines (SVMs) to train a classifier and then use the trained classifier to predict the class of new samples. SVMs are classifiers and can generally be used instead of Neural Networks. They are typically much faster to train than ANNs and don't require feature selection (filtering down to a small number of significant genes) to be performed.
Skills You Will Learn:
How to import a data set.
How to import a variable (class labels).
How to create, evaluate, and predict classes using a SVM committee classifier.
Scientific Background
This tutorial is a reanalysis of the data reported by Khan, Wei, Ringnér et al. in Nature Medicine (2001) [Ref.1]. We refer to this paper simply as 'Khan' in this tutorial.
The object of the paper and of this tutorial is to learn to distinguish, at the molecular level, between types of small round blue-cell tumors (SRBCTs) such as Ewing sarcoma (EWS), Burkitt lymphoma (BL), neuroblastoma (NB) and rhabdomyosarcoma (RMS). These tumors are difficult to distinguish by visual methods, and respond to different treatments.
The data is available on the World Wide Web as supplementary material, at http://www.thep.lu.se/pub/Preprints/01/lu_tp_01_06_supp.html. The authors pre-filtered the data for a minimal level of expression, leaving measurements for 2308 genes.
SVM Classifiers
SVMs are a type of machine learning algorithm that were invented by Vapnik. [4] They have been successfully applied to a wide range of pattern recognition and classification problems including handwriting recognition, face detection, and microarray gene expression analysis. Compared with artificial neural networks (ANNs) they are faster, can be used with larger numbers of genes, are more interpretable, and are deterministic. They find an optimal separating hyperplane between data points of different classes in a (possibly) high dimensional space. The actual Support Vectors are the points that form the decision boundary between the classes. Here is a simple example in 2D where a user is trying to separate data of two classes of samples (red circles and green squares):
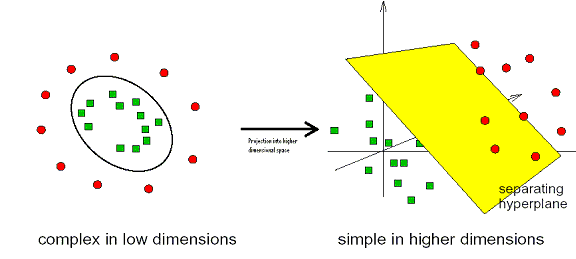 [3]
[3]
One of the main advantages of SVMs is that they are maximal margin classifiers. For example, in the plots below, D is a better separator than A, B or C since it will be more likely to classify new samples that are close to the current decision boundary:
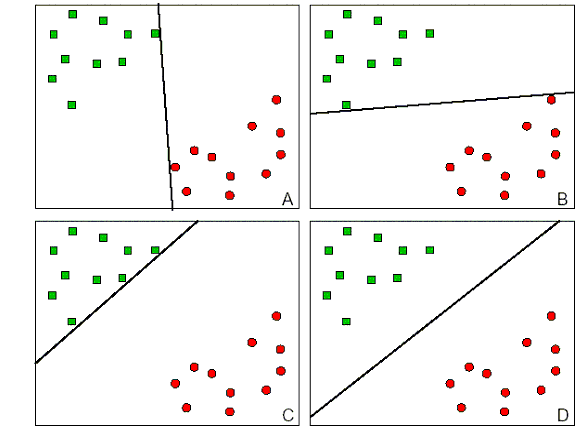 [3]
[3]
Because a single learner (of any type) can produce good or bad results on a particular data set, having a committee architecture improves the reliability of classification. Typically 10 is a reasonable number of committee members, with the requirement that 70% of committee members agree for a classification to be made.
Tutorial Workflow
The purpose of the workflow covered by this tutorial is to train a committee of SVMs to distinguish between different types of tumors. SVM training takes place on the training dataset. The samples in this dataset have known classes, so the SVM training is done under the supervision of this available knowledge. Once the SVM committee has been trained, it can be used to obtain a diagnosis for new samples of the same phenomenon (SRBCTs), to predict the classes of its samples. This new data is called the test dataset.
What You Will Learn:
1. How to train a committee of support vector machines (SVMs) .
2. How to use trained SVMs to distinguish and predict sample classes.
Tutorial Length
This tutorial should take about an hour, depending on how long you spend investigating the data, and how fast your machine is.
If you must stop part way through the tutorial, simply exit the program by selecting Exit from the File menu. The data and experiments you have performed to that point are saved automatically by GeneLinker™. The next time you start GeneLinker™, you can continue on with the next step in the tutorial.
This tutorial uses the same datasets and variables as Tutorial 6. If you have already performed tutorial 6, you can safely skip the data import and variable import steps and use the datasets that are already in your experiment navigator.

